Diffusion Equations
We start by writing a specialised solver for solving diffusion equations. What we produce in this section can also be accessed in FiniteVolumeMethod.DiffusionEquation.
Mathematical Details
Let us start by considering the mathematical details. The equation we consider is
\[\begin{equation} \begin{aligned} \pdv{u}{t} &= \div\left[D(\vb x)\grad u\right] & \vb x \in \Omega. \end{aligned} \end{equation}\]
From the mathematical details section (where we also define the notation that follows), we know that discretising this problem leads to an equation of the form
\[\dv{u_i}{t} + \frac{1}{V_i}\sum_{\sigma\in\mathcal E_i}\left[\vb q\left(\vb x_\sigma, t, \alpha_{k(\sigma)}x_\sigma+\beta_{k(\sigma)}y_\sigma+\gamma_{k(\sigma)}\right)\vdot\vu n\right]L_\sigma = S_i,\]
For the diffusion equation, the flux function is $\vb q = -D\grad u$, meaning for an interior node we have
\[\vb q(\vb x_\sigma, t, \alpha_{k(\sigma)}x_\sigma+\beta_{k(\sigma)}y_\sigma+\gamma_{k(\sigma)}) = -D(\vb x_\sigma)(\alpha_{k(\sigma)}, \beta_{k(\sigma)})^{\mkern-1.5mu\mathsf{T}}.\]
Thus, also using $S_i=0$,
\[\dv{u_i}{t} = \frac{1}{V_i}\sum_{\sigma\in\mathcal E_i} D(\vb x_\sigma)\left[\alpha_{k(\sigma)}n_\sigma^x + \beta_{k(\sigma)}n_\sigma^y\right]L_\sigma,\]
where $\vu n = (n_\sigma^x, n_\sigma^y)^{\mkern-1.5mu\mathsf{T}}$. It is still not immediately obvious how we can turn this into a linear problem. To see the linearity, note that
\[\begin{equation} \begin{aligned} \alpha_{k(\sigma)} = s_{k(\sigma), 11}u_{k(\sigma)1} + s_{k(\sigma), 12}u_{k(\sigma)2} + s_{k(\sigma), 13}u_{k(\sigma)3}, \\ \beta_{k(\sigma)} = s_{k(\sigma), 21}u_{k(\sigma)1} + s_{k(\sigma), 22}u_{k(\sigma)2} + s_{k(\sigma),23}u_{k(\sigma)3}, \\ \end{aligned} \end{equation}\]
thus, now writing $k=k(\sigma)$ for simplicity,
\[\begin{equation*} \begin{aligned} \dv{u_i}{t} &= \frac{1}{V_i}\sum_{\sigma\in \mathcal E_i} D(\vb x_\sigma)\left[\left(s_{k, 11}u_{k1} + s_{k, 12}u_{k2} + s_{k,13}u_{k3}\right)n_\sigma^x + \left(s_{k, 21}u_{k1} + s_{k, 22}u_{k2} + s_{k, 23}u_{k3}\right)n_\sigma^y\right]L_\sigma \\ &= \frac{1}{V_i}\sum_{\sigma\in\mathcal E_i} D(\vb x_\sigma)\left[\left(s_{k, 11}n_\sigma^x + s_{k, 21}n_\sigma^y\right)u_{k1} + \left(s_{k, 12}n_\sigma^x + s_{k, 22}n_\sigma^y\right)u_{k2} + \left(s_{k, 13}n_\sigma^x + s_{k, 23}n_\sigma^y\right)u_{k3}\right]L_\sigma \\ &= \vb a_i^{\mkern-1.5mu\mathsf{T}}\vb u, \end{aligned} \end{equation*}\]
Now, the result
\[\begin{equation}\label{eq:disc1} \dv{u_i}{t} = \vb a_i^{\mkern-1.5mu\mathsf{T}}\vb u + b_i, \end{equation}\]
where $b_i=0$, is for the case that $i$ is an interior node. We need to think about how boundary conditions get incorporated. For this problem, we will not allow the boundary conditions to depend on $u$ or $t$.[1]
Let's think about what we each type of boundary condition would to our problem.
- For a Dirichlet boundary condition, we have $u_i = a(\vb x_i)$ for some $\vb x_i$. To implement this, we let the $i$th row of $\vb A$ be zero and $b_i=0$. Then, as long as we start the Dirichlet nodes at $u_i=a(\vb x_i)$, they will stay at that value as $u_i' = 0$ there.[2]
- Suppose we have a Neumann boundary condition, say $\grad u \vdot \vu n = a(\vb x)$, we need to write the sum over $\sigma \in \mathcal E_i$ so that the differences between the boundary edges and the interior edges are made explicit. Over these boundary edges, we get sums that go into $\vb b$ rather than into $\vb A$.
- For conditions of the form $\mathrm du_i/\mathrm dt = a(\vb x_i)$, we should just set $\vb a_i = \vb 0$ and $b_i = a(\vb x_i)$. Note that here $\vb A$ is singular.
Implementation
We now know enough to implement our solver. Let us walk through this slowly, defining our function and then iterating it slowly to incorporate different features. The function signature will be similar to how we define an FVMProblem, namely
function diffusion_equation(mesh::FVMGeometry,
BCs::BoundaryConditions,
ICs::InternalConditions=InternalConditions();
diffusion_function,
diffusion_parameters=nothing,
initial_condition,
initial_time=0.0,
final_time)
# return the ODEProblem
endFor the boundary and internal conditions, we'll assume that the functions take the same form, i.e. (x, y, t, u, p) -> Number, but the t and u arguments will both be passed as nothing. The diffusion function should be of the form (x, y, p) -> Number, or simply a Number.
We need to first write a function that will construct $(\vb A, \vb b)$. The idea for this is to loop over each triangle and then pick up the contributions, and then over all the boundary edges, just as we describe in the mathematical details section. The main difference being that, rather than adding terms to $\mathrm du_i/\mathrm dt$, we are picking out terms for $b_i$ and also to put into $\vb A$.
Let us start by writing out the contribution from all the triangles.
using FiniteVolumeMethod
const FVM = FiniteVolumeMethod
function triangle_contributions!(A, mesh, conditions, diffusion_function, diffusion_parameters)
for T in each_solid_triangle(mesh.triangulation)
ijk = triangle_vertices(T)
i, j, k = ijk
props = FVM.get_triangle_props(mesh, i, j, k)
s₁₁, s₁₂, s₁₃, s₂₁, s₂₂, s₂₃, s₃₁, s₃₂, s₃₃ = props.shape_function_coefficients
for (edge_index, (e1, e2)) in enumerate(((i, j), (j, k), (k, i)))
x, y, nx, ny, ℓ = FVM.get_cv_components(props, edge_index)
D = diffusion_function(x, y, diffusion_parameters)
Dℓ = D * ℓ
a123 = (Dℓ * (s₁₁ * nx + s₂₁ * ny),
Dℓ * (s₁₂ * nx + s₂₂ * ny),
Dℓ * (s₁₃ * nx + s₂₃ * ny))
e1_hascond = FVM.has_condition(conditions, e1)
e2_hascond = FVM.has_condition(conditions, e2)
for vert in 1:3
e1_hascond || (A[e1, ijk[vert]] += a123[vert] / FVM.get_volume(mesh, e1))
e2_hascond || (A[e2, ijk[vert]] -= a123[vert] / FVM.get_volume(mesh, e2))
end
end
end
endtriangle_contributions! (generic function with 1 method)Now we need the function that gets the contributions from the boundary edges.
function boundary_edge_contributions!(A, b, mesh, conditions,
diffusion_function, diffusion_parameters)
for e in keys(get_boundary_edge_map(mesh.triangulation))
i, j = DelaunayTriangulation.edge_vertices(e)
nx, ny, mᵢx, mᵢy, mⱼx, mⱼy, ℓ, T, props = FVM.get_boundary_cv_components(mesh, i, j)
ijk = triangle_vertices(T)
s₁₁, s₁₂, s₁₃, s₂₁, s₂₂, s₂₃, s₃₁, s₃₂, s₃₃ = props.shape_function_coefficients
Dᵢ = diffusion_function(mᵢx, mᵢy, diffusion_parameters)
Dⱼ = diffusion_function(mⱼx, mⱼy, diffusion_parameters)
i_hascond = FVM.has_condition(conditions, i)
j_hascond = FVM.has_condition(conditions, j)
if FVM.is_neumann_edge(conditions, i, j)
fidx = FVM.get_neumann_fidx(conditions, i, j)
aᵢ = FVM.eval_condition_fnc(conditions, fidx, mᵢx, mᵢy, nothing, nothing)
aⱼ = FVM.eval_condition_fnc(conditions, fidx, mⱼx, mⱼy, nothing, nothing)
i_hascond || (b[i] += Dᵢ * aᵢ * ℓ / FVM.get_volume(mesh, i))
j_hascond || (b[j] += Dⱼ * aⱼ * ℓ / FVM.get_volume(mesh, j))
else
aᵢ123 = (Dᵢ * ℓ * (s₁₁ * nx + s₂₁ * ny),
Dᵢ * ℓ * (s₁₂ * nx + s₂₂ * ny),
Dᵢ * ℓ * (s₁₃ * nx + s₂₃ * ny))
aⱼ123 = (Dⱼ * ℓ * (s₁₁ * nx + s₂₁ * ny),
Dⱼ * ℓ * (s₁₂ * nx + s₂₂ * ny),
Dⱼ * ℓ * (s₁₃ * nx + s₂₃ * ny))
for vert in 1:3
i_hascond || (A[i, ijk[vert]] += aᵢ123[vert] / FVM.get_volume(mesh, i))
j_hascond || (A[j, ijk[vert]] += aⱼ123[vert] / FVM.get_volume(mesh, i))
end
end
end
endboundary_edge_contributions! (generic function with 1 method)Now that we have the parts for handling the main flux contributions, we need to consider the boundary conditions. Note that in the code above we have alredy taken not to update $\vb A$ or $\vb b$ if there a boundary condition at the associated node, so we do not need to worry about e.g. zeroing out rows of $\vb A$ for a node with a boundary condition.
function apply_dirichlet_conditions!(initial_condition, mesh, conditions)
for (i, function_index) in FVM.get_dirichlet_nodes(conditions)
x, y = get_point(mesh, i)
initial_condition[i] = FVM.eval_condition_fnc(conditions, function_index, x, y, nothing, nothing)
end
end
function apply_dudt_conditions!(b, mesh, conditions)
for (i, function_index) in FVM.get_dudt_nodes(conditions)
if !FVM.is_dirichlet_node(conditions, i) # overlapping edges can be both Dudt and Dirichlet. Dirichlet takes precedence
x, y = get_point(mesh, i)
b[i] = FVM.eval_condition_fnc(conditions, function_index, x, y, nothing, nothing)
end
end
endapply_dudt_conditions! (generic function with 1 method)Now let's define diffusion_equation. For this, we note we want to write the problem in the form
\[\dv{\vb u}{t} = \vb A\vb u\]
to get the most out of our linearity in OrdinaryDiffEq.jl, whereas we currently have
\[\dv{\vb u}{t} = \vb A\vb u + \vb b.\]
To get around this, we define
\[\tilde{\vb u} = \begin{bmatrix} \vb u \\ 1 \end{bmatrix}, \quad \tilde{\vb A} = \begin{bmatrix}\vb A & \vb b \\ \vb 0^{\mkern-1.5mu\mathsf{T}} & 0 \end{bmatrix},\]
so that
\[\dv{\tilde{\vb u}}{t} = \begin{bmatrix} \vb u' \\ 0 \end{bmatrix} = \begin{bmatrix} \vb A\vb u + \vb b \\ 0 \end{bmatrix} = \tilde{\vb A}\tilde{\vb u}.\]
Note that this also requires that we append a 1 to the initial condition.
function diffusion_equation(mesh::FVMGeometry,
BCs::BoundaryConditions,
ICs::InternalConditions=InternalConditions();
diffusion_function,
diffusion_parameters=nothing,
initial_condition,
initial_time=0.0,
final_time)
conditions = Conditions(mesh, BCs, ICs)
n = DelaunayTriangulation.num_solid_vertices(mesh.triangulation)
Afull = zeros(n + 1, n + 1)
A = @views Afull[begin:end-1, begin:end-1]
b = @views Afull[begin:end-1, end]
_ic = vcat(initial_condition, 1)
triangle_contributions!(A, mesh, conditions, diffusion_function, diffusion_parameters)
boundary_edge_contributions!(A, b, mesh, conditions, diffusion_function, diffusion_parameters)
apply_dudt_conditions!(b, mesh, conditions)
apply_dirichlet_conditions!(_ic, mesh, conditions)
A_op = MatrixOperator(sparse(Afull))
prob = ODEProblem(A_op, _ic, (initial_time, final_time))
return prob
enddiffusion_equation (generic function with 2 methods)Let's now test the function. We use the same problem as in this tutorial.
using DelaunayTriangulation, OrdinaryDiffEq, LinearAlgebra, SparseArrays
tri = triangulate_rectangle(0, 2, 0, 2, 50, 50, single_boundary=true)
mesh = FVMGeometry(tri)
BCs = BoundaryConditions(mesh, (x, y, t, u, p) -> zero(x), Dirichlet)
diffusion_function = (x, y, p) -> 1 / 9
initial_condition = [y ≤ 1.0 ? 50.0 : 0.0 for (x, y) in DelaunayTriangulation.each_point(tri)]
final_time = 0.5
prob = diffusion_equation(mesh, BCs;
diffusion_function,
initial_condition,
final_time)
sol = solve(prob, Tsit5(); saveat=0.05)retcode: Success
Interpolation: 1st order linear
t: 11-element Vector{Float64}:
0.0
0.05
0.1
0.15
0.2
0.25
0.3
0.35
0.4
0.45
0.5
u: 11-element Vector{Vector{Float64}}:
[0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0 … 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 1.0]
[0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0 … 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 1.0]
[0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0 … 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 1.0]
[0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0 … 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 1.0]
[0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0 … 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 1.0]
[0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0 … 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 1.0]
[0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0 … 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 1.0]
[0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0 … 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 1.0]
[0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0 … 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 1.0]
[0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0 … 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 1.0]
[0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0 … 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 1.0](It would be nice to use LinearExponential() in the call above, but it just seems to be extremely numerically unstable, so it's unusable.) Note also that sol contains an extra component:
length(sol.u[1])2501DelaunayTriangulation.num_solid_vertices(tri)2500This is because we needed to add in an extra component to represent the problem as a linear problem. So, the solution is in sol[begin:end-1, :], and you should ignore sol[end, :]. (The same applies to DiffusionEquation that we introduce later.)
Let's now plot.
using CairoMakie
fig = Figure(fontsize=38)
for (i, j) in zip(1:3, (1, 6, 11))
ax = Axis(fig[1, i], width=600, height=600,
xlabel="x", ylabel="y",
title="t = $(sol.t[j])",
titlealign=:left)
u = j == 1 ? initial_condition : sol.u[j] # sol.u[1] is modified slightly to force the Dirichlet conditions at t = 0
tricontourf!(ax, tri, u, levels=0:5:50, colormap=:matter, extendlow=:auto, extendhigh=:auto) # don't need to do u[begin:end-1], since tri doesn't have that extra vertex.
tightlimits!(ax)
end
resize_to_layout!(fig)
fig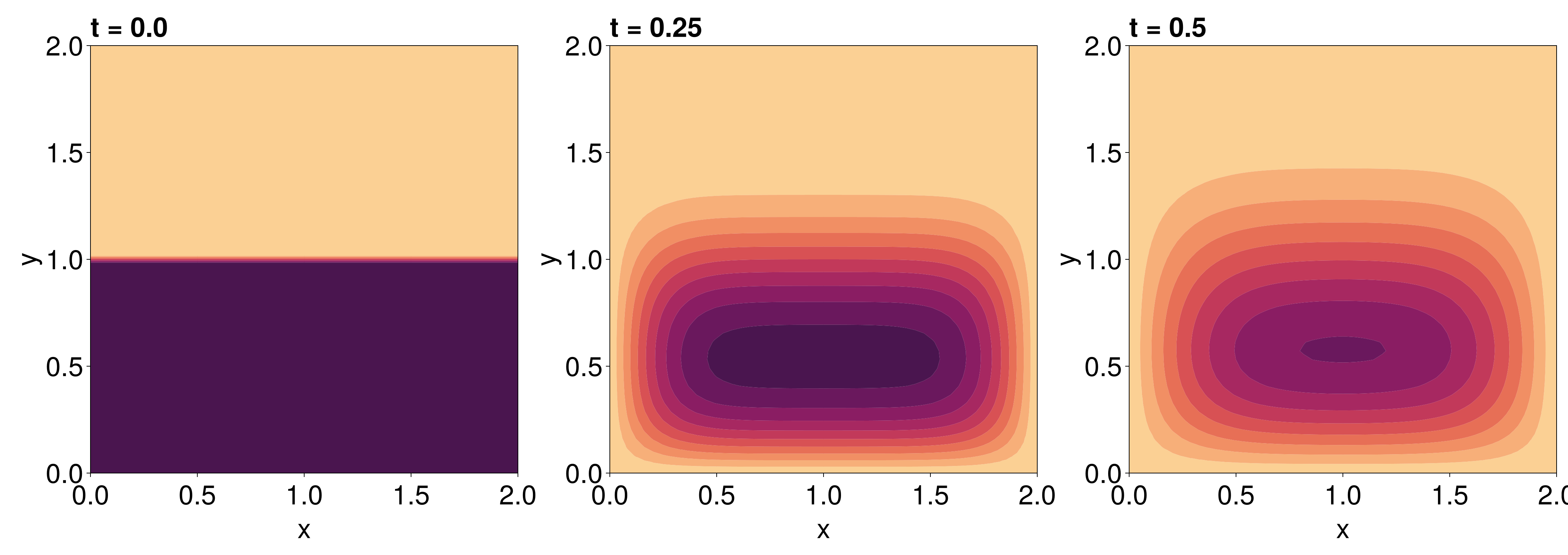
This is exactly the solution we expect!
Using the Provided Template
Let's now use the built-in DiffusionEquation() which implements the above template inside FiniteVolumeMethod.jl.
diff_eq = DiffusionEquation(mesh, BCs;
diffusion_function,
initial_condition,
final_time)DiffusionEquation with 2500 nodes and time span (0.0, 0.5)Let's compare DiffusionEquation to the FVMProblem approach.
fvm_prob = FVMProblem(mesh, BCs;
diffusion_function=let D = diffusion_function
(x, y, t, u, p) -> D(x, y, p)
end,
initial_condition,
final_time)FVMProblem with 2500 nodes and time span (0.0, 0.5)using BenchmarkTools
@btime solve($diff_eq, $Tsit5(), saveat=$0.05); 5.736 ms (82 allocations: 552.42 KiB)using LinearSolve
@btime solve($fvm_prob, $TRBDF2(linsolve=KLUFactorization()), saveat=$0.05); 49.237 ms (91755 allocations: 32.02 MiB)Much better! The DiffusionEquation approach is about 10 times faster.
To finish this example, let's solve a diffusion equation with constant Neumann boundary conditions:
\[\begin{equation*} \begin{aligned} \pdv{u}{t} &= 2\grad^2 u & \vb x \in \Omega, \\ \grad u \vdot \vu n &= 2 & \vb x \in \partial\Omega. \end{aligned} \end{equation*}\]
Here, $\Omega = [0, 320]^2$.
L = 320.0
tri = triangulate_rectangle(0, L, 0, L, 100, 100, single_boundary=true)
mesh = FVMGeometry(tri)
BCs = BoundaryConditions(mesh, (x, y, t, u, p) -> 2.0, Neumann)
diffusion_function = (x, y, p) -> 2.0
initf = (x, y) -> begin
if 0.4L ≤ y ≤ 0.6L
return 1.0
else
return 0.0
end
end
final_time = 500.0
initial_condition = [initf(x, y) for (x, y) in DelaunayTriangulation.each_point(tri)]
prob = DiffusionEquation(mesh, BCs;
diffusion_function,
initial_condition,
final_time)DiffusionEquation with 10000 nodes and time span (0.0, 500.0)Let's solve and plot.
sol = solve(prob, Tsit5(); saveat=100.0)retcode: Success
Interpolation: 1st order linear
t: 6-element Vector{Float64}:
0.0
100.0
200.0
300.0
400.0
500.0
u: 6-element Vector{Vector{Float64}}:
[0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0 … 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 1.0]
[63.69432146948662, 57.91993563851779, 52.468254485047616, 48.07467536264921, 44.32409891376271, 41.312319386306044, 38.87775229689681, 36.98180247606284, 35.51874424288336, 34.42222343912825 … 35.51874424287919, 36.9818024760664, 38.87775229688911, 41.312319386320816, 44.324098913738624, 48.07467536269855, 52.46825448494263, 57.91993563875677, 63.69432146886061, 1.0]
[91.50473260688723, 83.71760875996453, 78.69493124267801, 73.3849378927463, 69.04806689875537, 65.04397421701678, 61.648665579203154, 58.66588690809526, 56.14262845370988, 53.99225264855068 … 56.142628453714934, 58.66588690808136, 61.64866557922738, 65.04397421697105, 69.04806689884693, 73.38493789256844, 78.69493124304407, 83.71760875913677, 91.5047326090407, 1.0]
[111.3264796893097, 104.12803651686592, 98.70364195388714, 93.24912184952623, 88.50484048430158, 84.09374545497968, 80.1785823663648, 76.64390606135497, 73.51698409166313, 70.74569558372897 … 73.51698409166475, 76.64390606134724, 80.17858236637763, 84.09374545495513, 88.5048404843528, 93.24912184942808, 98.70364195408713, 104.12803651641441, 111.32647969048199, 1.0]
[125.84898878011737, 122.18663715509418, 115.25163624904545, 110.28509663472774, 105.01236092340481, 100.49453351161048, 96.1788470217854, 92.33274745412113, 88.7904377122623, 85.61578063131344 … 88.79043771224866, 92.33274745414077, 96.1788470217473, 100.49453351168056, 105.01236092327554, 110.28509663498563, 115.25163624850686, 122.18663715631531, 125.84898877693128, 1.0]
[141.75921595314702, 136.91169589100613, 130.381410153159, 125.09716189470845, 119.78045994667951, 115.02374593979808, 110.51491577596983, 106.40139024536721, 102.58742470687305, 99.10565520758897 … 102.58742470686482, 106.40139024537704, 110.51491577595006, 115.0237459398341, 119.78045994661478, 125.09716189483846, 130.38141015288542, 136.9116958916271, 141.75921595152502, 1.0]fig = Figure(fontsize=38)
for j in eachindex(sol)
ax = Axis(fig[1, j], width=600, height=600,
xlabel="x", ylabel="y",
title="t = $(sol.t[j])",
titlealign=:left)
u = j == 1 ? initial_condition : sol.u[j]
tricontourf!(ax, tri, u, levels=0:0.1:1, colormap=:turbo, extendlow=:auto, extendhigh=:auto)
tightlimits!(ax)
end
resize_to_layout!(fig)
fig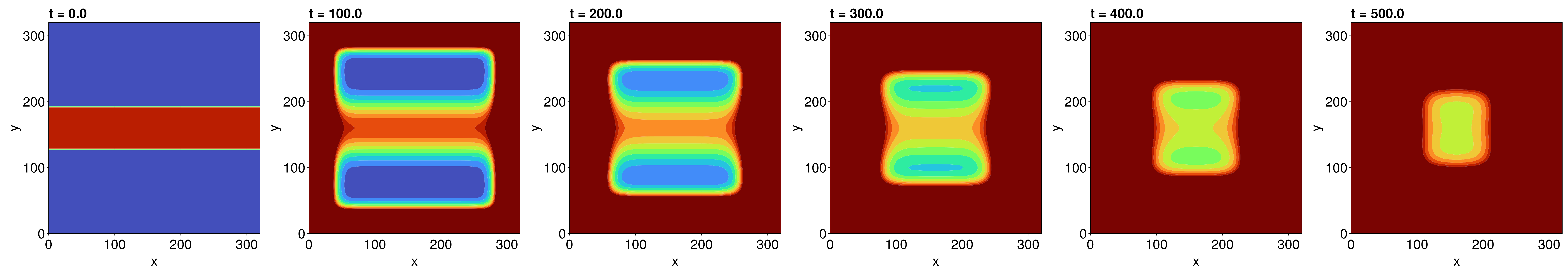
For the corresponding FVMProblem, note that the Neumann boundary conditions need to be defined in terms of $\vb q = -D(\vb x)\grad u$ rather than $\grad u \vdot \vu n$. So, since $\grad u \vdot \vu n = 2$, we have $-D\grad u \vdot \vu n = -2D = -4$, so $\vb q \vdot \vu n = -4$. Here is a comparison of the two solutions.
BCs_prob = BoundaryConditions(mesh, (x, y, t, u, p) -> -4, Neumann)
fvm_prob = FVMProblem(mesh, BCs_prob;
diffusion_function=let D = diffusion_function
(x, y, t, u, p) -> D(x, y, p)
end,
initial_condition,
final_time)
fvm_sol = solve(fvm_prob, TRBDF2(linsolve=KLUFactorization()); saveat=100.0)
fvm_sol |> tc #hide
for j in eachindex(fvm_sol)
ax = Axis(fig[2, j], width=600, height=600,
xlabel="x", ylabel="y",
title="t = $(fvm_sol.t[j])",
titlealign=:left)
u = j == 1 ? initial_condition : fvm_sol.u[j]
tricontourf!(ax, tri, u, levels=0:0.1:1, colormap=:turbo, extendlow=:auto, extendhigh=:auto)
tightlimits!(ax)
end
resize_to_layout!(fig)
figHere is a benchmark comparison.
@btime solve($prob, $Tsit5(), saveat=$100.0); 78.761 ms (71 allocations: 1.76 MiB)using Sundials
@btime solve($fvm_prob, $CVODE_BDF(linear_solver=:GMRES), saveat=$100.0); 94.839 ms (111666 allocations: 56.07 MiB)These problems also work with the pl_interpolate function:
q = (30.0, 45.0)
T = jump_and_march(tri, q)
val = pl_interpolate(prob, T, sol.u[3], q[1], q[2])11.090458780694902Just the code
An uncommented version of this example is given below. You can view the source code for this file here.
using FiniteVolumeMethod
const FVM = FiniteVolumeMethod
function triangle_contributions!(A, mesh, conditions, diffusion_function, diffusion_parameters)
for T in each_solid_triangle(mesh.triangulation)
ijk = triangle_vertices(T)
i, j, k = ijk
props = FVM.get_triangle_props(mesh, i, j, k)
s₁₁, s₁₂, s₁₃, s₂₁, s₂₂, s₂₃, s₃₁, s₃₂, s₃₃ = props.shape_function_coefficients
for (edge_index, (e1, e2)) in enumerate(((i, j), (j, k), (k, i)))
x, y, nx, ny, ℓ = FVM.get_cv_components(props, edge_index)
D = diffusion_function(x, y, diffusion_parameters)
Dℓ = D * ℓ
a123 = (Dℓ * (s₁₁ * nx + s₂₁ * ny),
Dℓ * (s₁₂ * nx + s₂₂ * ny),
Dℓ * (s₁₃ * nx + s₂₃ * ny))
e1_hascond = FVM.has_condition(conditions, e1)
e2_hascond = FVM.has_condition(conditions, e2)
for vert in 1:3
e1_hascond || (A[e1, ijk[vert]] += a123[vert] / FVM.get_volume(mesh, e1))
e2_hascond || (A[e2, ijk[vert]] -= a123[vert] / FVM.get_volume(mesh, e2))
end
end
end
end
function boundary_edge_contributions!(A, b, mesh, conditions,
diffusion_function, diffusion_parameters)
for e in keys(get_boundary_edge_map(mesh.triangulation))
i, j = DelaunayTriangulation.edge_vertices(e)
nx, ny, mᵢx, mᵢy, mⱼx, mⱼy, ℓ, T, props = FVM.get_boundary_cv_components(mesh, i, j)
ijk = triangle_vertices(T)
s₁₁, s₁₂, s₁₃, s₂₁, s₂₂, s₂₃, s₃₁, s₃₂, s₃₃ = props.shape_function_coefficients
Dᵢ = diffusion_function(mᵢx, mᵢy, diffusion_parameters)
Dⱼ = diffusion_function(mⱼx, mⱼy, diffusion_parameters)
i_hascond = FVM.has_condition(conditions, i)
j_hascond = FVM.has_condition(conditions, j)
if FVM.is_neumann_edge(conditions, i, j)
fidx = FVM.get_neumann_fidx(conditions, i, j)
aᵢ = FVM.eval_condition_fnc(conditions, fidx, mᵢx, mᵢy, nothing, nothing)
aⱼ = FVM.eval_condition_fnc(conditions, fidx, mⱼx, mⱼy, nothing, nothing)
i_hascond || (b[i] += Dᵢ * aᵢ * ℓ / FVM.get_volume(mesh, i))
j_hascond || (b[j] += Dⱼ * aⱼ * ℓ / FVM.get_volume(mesh, j))
else
aᵢ123 = (Dᵢ * ℓ * (s₁₁ * nx + s₂₁ * ny),
Dᵢ * ℓ * (s₁₂ * nx + s₂₂ * ny),
Dᵢ * ℓ * (s₁₃ * nx + s₂₃ * ny))
aⱼ123 = (Dⱼ * ℓ * (s₁₁ * nx + s₂₁ * ny),
Dⱼ * ℓ * (s₁₂ * nx + s₂₂ * ny),
Dⱼ * ℓ * (s₁₃ * nx + s₂₃ * ny))
for vert in 1:3
i_hascond || (A[i, ijk[vert]] += aᵢ123[vert] / FVM.get_volume(mesh, i))
j_hascond || (A[j, ijk[vert]] += aⱼ123[vert] / FVM.get_volume(mesh, i))
end
end
end
end
function apply_dirichlet_conditions!(initial_condition, mesh, conditions)
for (i, function_index) in FVM.get_dirichlet_nodes(conditions)
x, y = get_point(mesh, i)
initial_condition[i] = FVM.eval_condition_fnc(conditions, function_index, x, y, nothing, nothing)
end
end
function apply_dudt_conditions!(b, mesh, conditions)
for (i, function_index) in FVM.get_dudt_nodes(conditions)
if !FVM.is_dirichlet_node(conditions, i) # overlapping edges can be both Dudt and Dirichlet. Dirichlet takes precedence
x, y = get_point(mesh, i)
b[i] = FVM.eval_condition_fnc(conditions, function_index, x, y, nothing, nothing)
end
end
end
function diffusion_equation(mesh::FVMGeometry,
BCs::BoundaryConditions,
ICs::InternalConditions=InternalConditions();
diffusion_function,
diffusion_parameters=nothing,
initial_condition,
initial_time=0.0,
final_time)
conditions = Conditions(mesh, BCs, ICs)
n = DelaunayTriangulation.num_solid_vertices(mesh.triangulation)
Afull = zeros(n + 1, n + 1)
A = @views Afull[begin:end-1, begin:end-1]
b = @views Afull[begin:end-1, end]
_ic = vcat(initial_condition, 1)
triangle_contributions!(A, mesh, conditions, diffusion_function, diffusion_parameters)
boundary_edge_contributions!(A, b, mesh, conditions, diffusion_function, diffusion_parameters)
apply_dudt_conditions!(b, mesh, conditions)
apply_dirichlet_conditions!(_ic, mesh, conditions)
A_op = MatrixOperator(sparse(Afull))
prob = ODEProblem(A_op, _ic, (initial_time, final_time))
return prob
end
using DelaunayTriangulation, OrdinaryDiffEq, LinearAlgebra, SparseArrays
tri = triangulate_rectangle(0, 2, 0, 2, 50, 50, single_boundary=true)
mesh = FVMGeometry(tri)
BCs = BoundaryConditions(mesh, (x, y, t, u, p) -> zero(x), Dirichlet)
diffusion_function = (x, y, p) -> 1 / 9
initial_condition = [y ≤ 1.0 ? 50.0 : 0.0 for (x, y) in DelaunayTriangulation.each_point(tri)]
final_time = 0.5
prob = diffusion_equation(mesh, BCs;
diffusion_function,
initial_condition,
final_time)
sol = solve(prob, Tsit5(); saveat=0.05)
length(sol.u[1])
DelaunayTriangulation.num_solid_vertices(tri)
using CairoMakie
fig = Figure(fontsize=38)
for (i, j) in zip(1:3, (1, 6, 11))
ax = Axis(fig[1, i], width=600, height=600,
xlabel="x", ylabel="y",
title="t = $(sol.t[j])",
titlealign=:left)
u = j == 1 ? initial_condition : sol.u[j] # sol.u[1] is modified slightly to force the Dirichlet conditions at t = 0
tricontourf!(ax, tri, u, levels=0:5:50, colormap=:matter, extendlow=:auto, extendhigh=:auto) # don't need to do u[begin:end-1], since tri doesn't have that extra vertex.
tightlimits!(ax)
end
resize_to_layout!(fig)
fig
diff_eq = DiffusionEquation(mesh, BCs;
diffusion_function,
initial_condition,
final_time)
fvm_prob = FVMProblem(mesh, BCs;
diffusion_function=let D = diffusion_function
(x, y, t, u, p) -> D(x, y, p)
end,
initial_condition,
final_time)
L = 320.0
tri = triangulate_rectangle(0, L, 0, L, 100, 100, single_boundary=true)
mesh = FVMGeometry(tri)
BCs = BoundaryConditions(mesh, (x, y, t, u, p) -> 2.0, Neumann)
diffusion_function = (x, y, p) -> 2.0
initf = (x, y) -> begin
if 0.4L ≤ y ≤ 0.6L
return 1.0
else
return 0.0
end
end
final_time = 500.0
initial_condition = [initf(x, y) for (x, y) in DelaunayTriangulation.each_point(tri)]
prob = DiffusionEquation(mesh, BCs;
diffusion_function,
initial_condition,
final_time)
sol = solve(prob, Tsit5(); saveat=100.0)
fig = Figure(fontsize=38)
for j in eachindex(sol)
ax = Axis(fig[1, j], width=600, height=600,
xlabel="x", ylabel="y",
title="t = $(sol.t[j])",
titlealign=:left)
u = j == 1 ? initial_condition : sol.u[j]
tricontourf!(ax, tri, u, levels=0:0.1:1, colormap=:turbo, extendlow=:auto, extendhigh=:auto)
tightlimits!(ax)
end
resize_to_layout!(fig)
fig
BCs_prob = BoundaryConditions(mesh, (x, y, t, u, p) -> -4, Neumann)
fvm_prob = FVMProblem(mesh, BCs_prob;
diffusion_function=let D = diffusion_function
(x, y, t, u, p) -> D(x, y, p)
end,
initial_condition,
final_time)
fvm_sol = solve(fvm_prob, TRBDF2(linsolve=KLUFactorization()); saveat=100.0)
for j in eachindex(fvm_sol)
ax = Axis(fig[2, j], width=600, height=600,
xlabel="x", ylabel="y",
title="t = $(fvm_sol.t[j])",
titlealign=:left)
u = j == 1 ? initial_condition : fvm_sol.u[j]
tricontourf!(ax, tri, u, levels=0:0.1:1, colormap=:turbo, extendlow=:auto, extendhigh=:auto)
tightlimits!(ax)
end
resize_to_layout!(fig)
fig
q = (30.0, 45.0)
T = jump_and_march(tri, q)
val = pl_interpolate(prob, T, sol.u[3], q[1], q[2])This page was generated using Literate.jl.
- 1It would be fine to allow the boundary conditions to depend on $t$ - we would still have linearity. The issue would just be that we need to reconstruct the matrix at every time step. So, for simplicity, let's not allow it so that the template we build is efficient for the most common case (where there is no $t$ dependence).
- 2If the boundary condition was non-autonomous, we could use a mass matrix instead, or build the condition into $\vb A$ and $\vb b$ directly by using the exact values of $u$ where applicable.